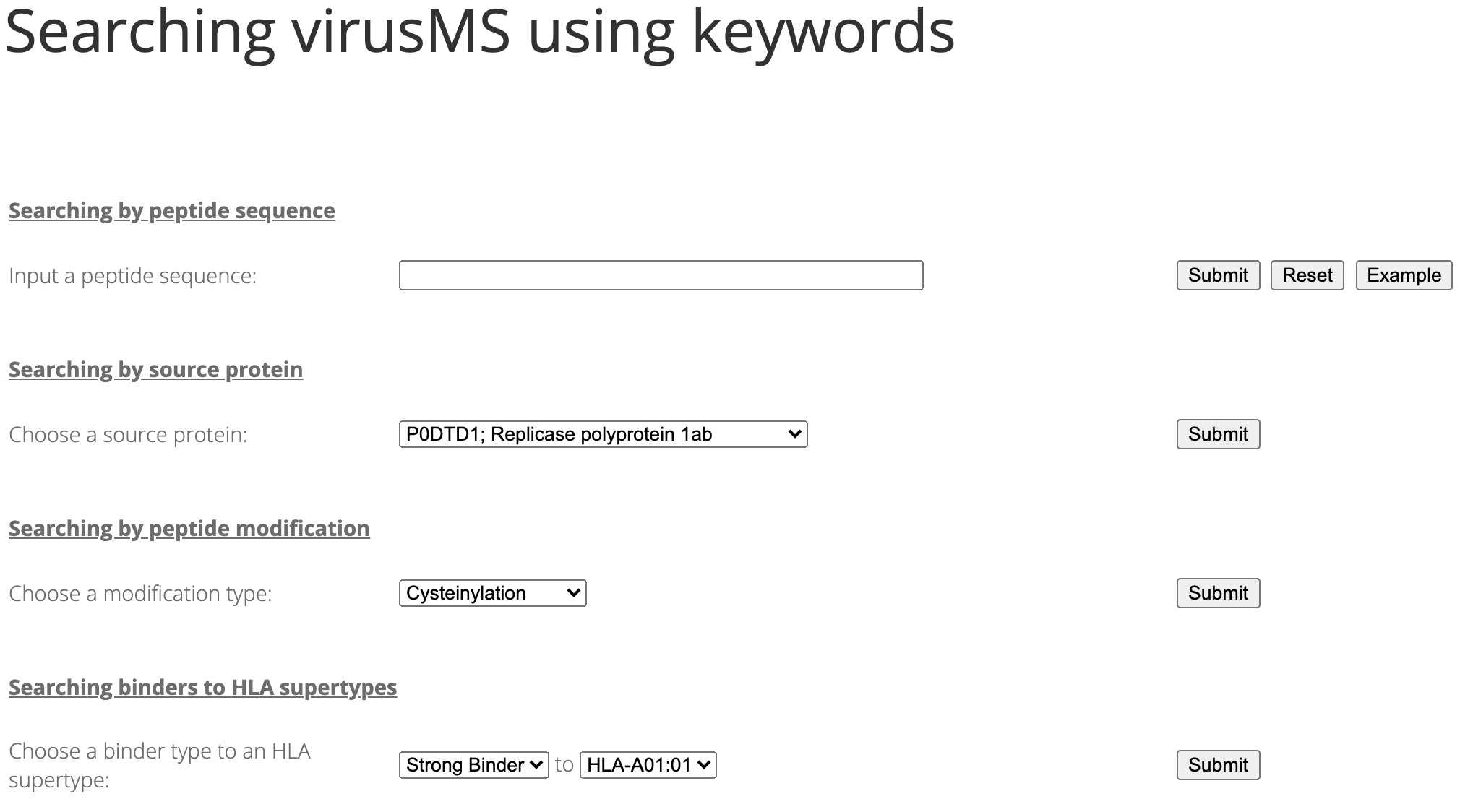
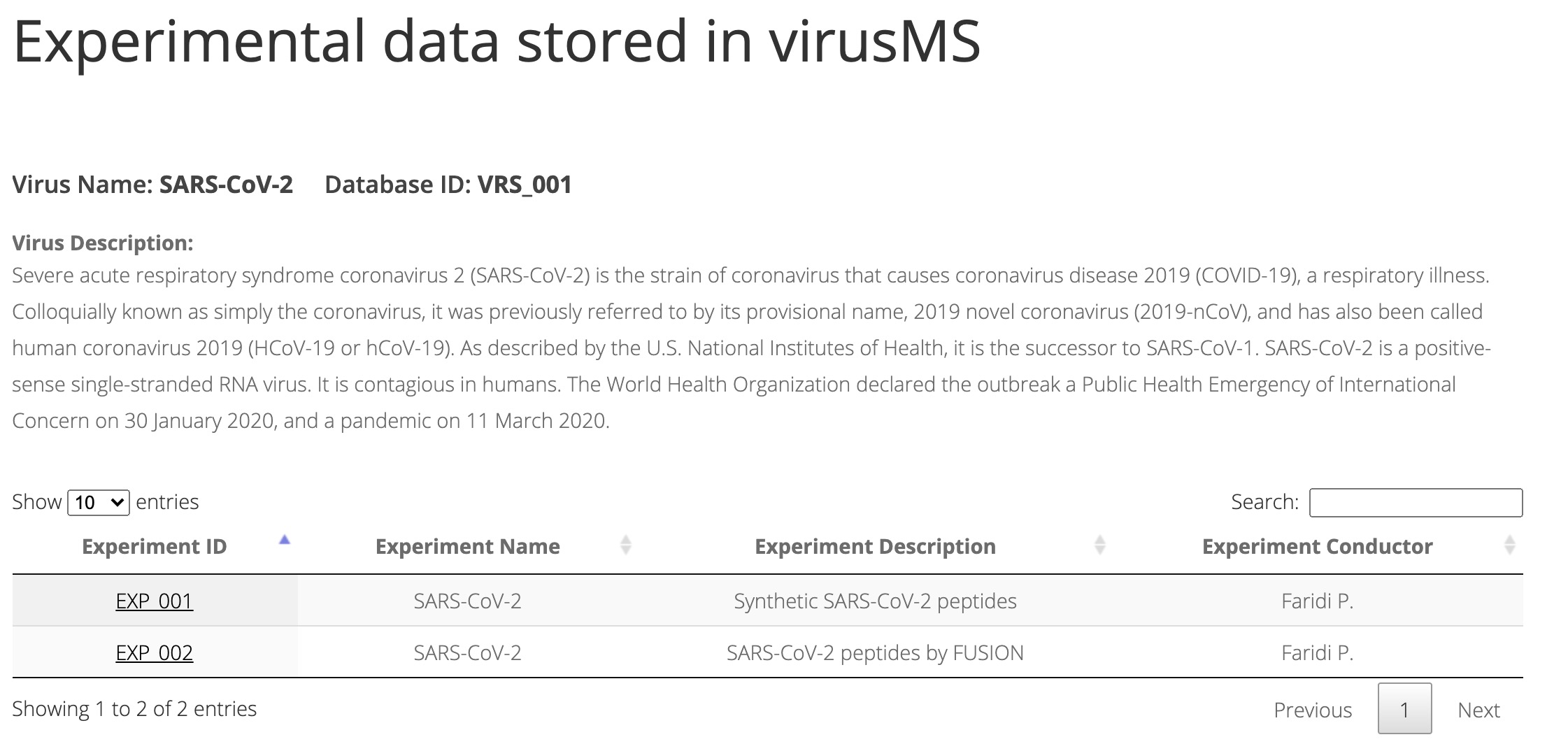
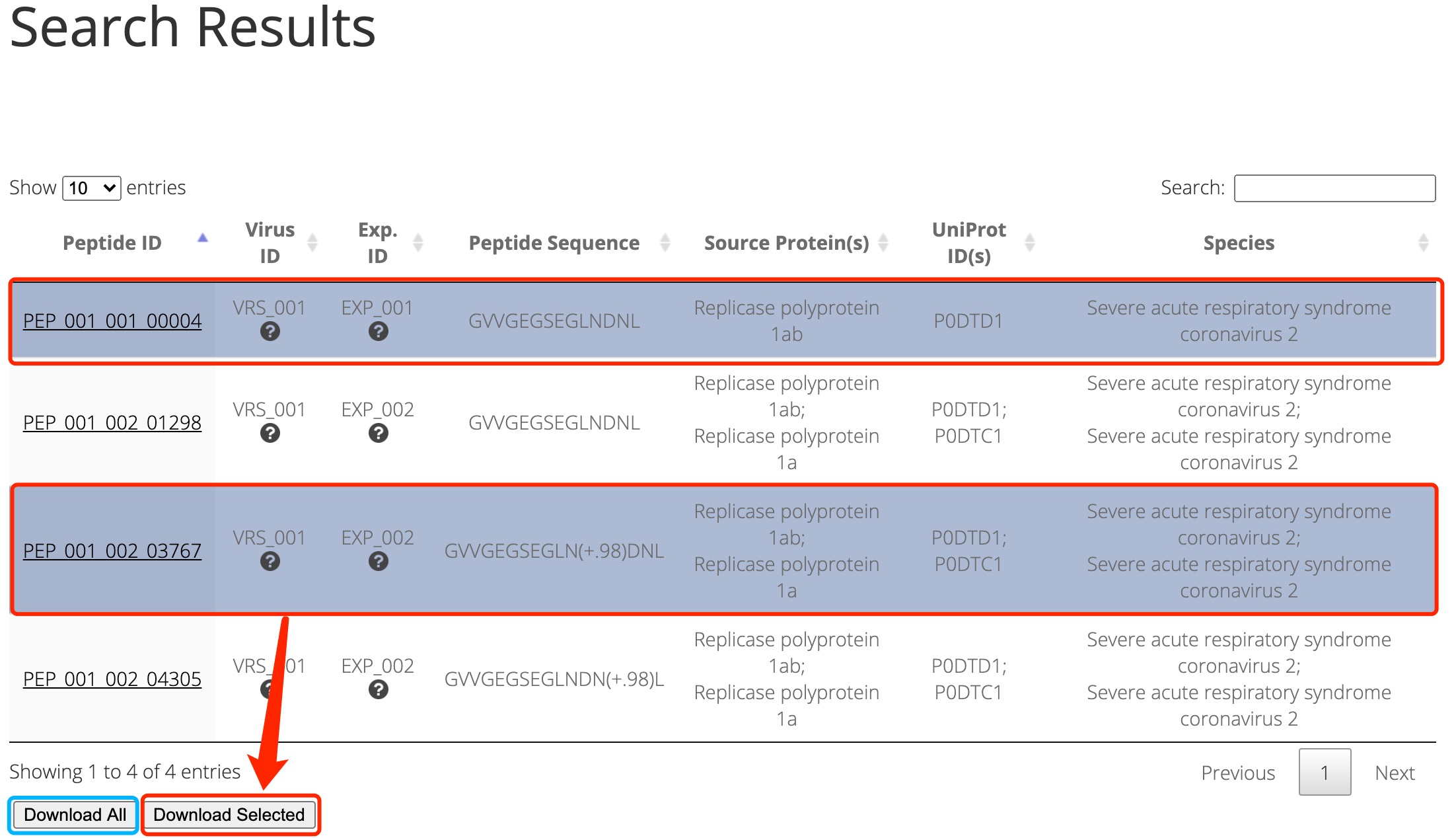
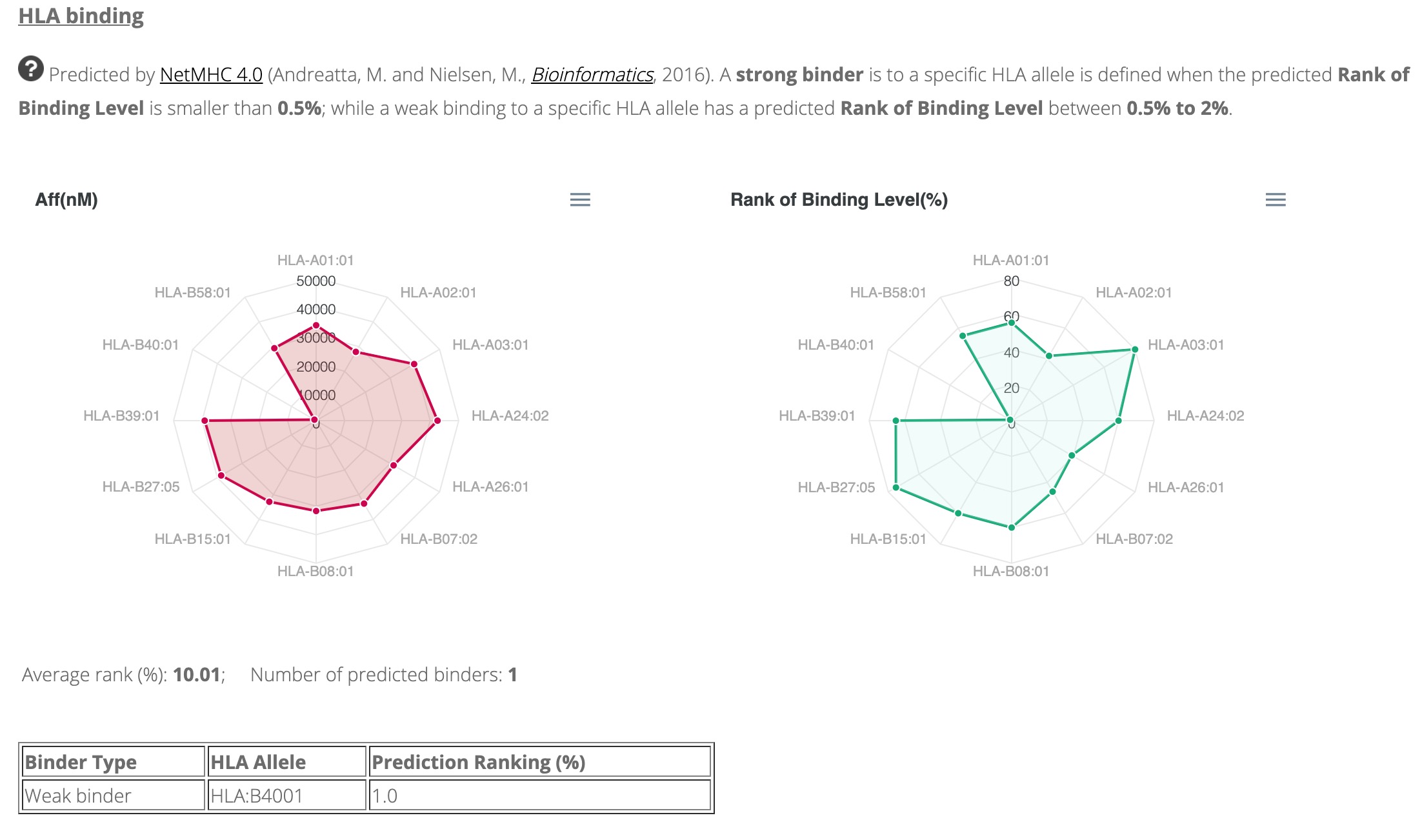
SARS-CoV-2 has caused a significant ongoing pandemic worldwide. A number of studies have examined the T cell mediated immune responses against SARS-CoV-2. Accurately identifying potential T cell epitopes derived from SARS-CoV-2 proteome will aid in identifying targets for vaccination. In this study, we applied tandem mass spectrometry and proteomic techniques to a library of ~40,000 synthetic peptides, in order to generate a large dataset of SARS-CoV-2 peptide spectra. On this basis, we built an online knowledgebase, termed virusMS, to document, annotate and analyse these synthetic peptides and their spectral information. VirusMS incorporates a user-friendly interface to facilitate searching, browsing and downloading the database content. Detailed annotations of the peptides, including experimental information, peptide modifications, predicted peptide-HLA (human leukocyte antigen) binding affinities, and peptide MS/MS spectra data, are provided in virusMS.
Citation
If you think viruMS is useful in your research, please consider citing us:
Acknowledgement

The authors acknowledge the donation of a peptide library spanning the entire sequence of the SARS-CoV-2 proteome by Mimotopes Pty Ltd (Mulgrave, Victoria, Australia).

Computational resources were supported by the R@CMon/Monash Node of the Nectar Research Cloud, an initiative of the Australian Government's Super Science Scheme and the Education Investment Fund.