The virusMS database is indexed by the virus and experiment. The Browse function will bring you the page where all viruses and their corresponding experiments will be shown (Figure 1).
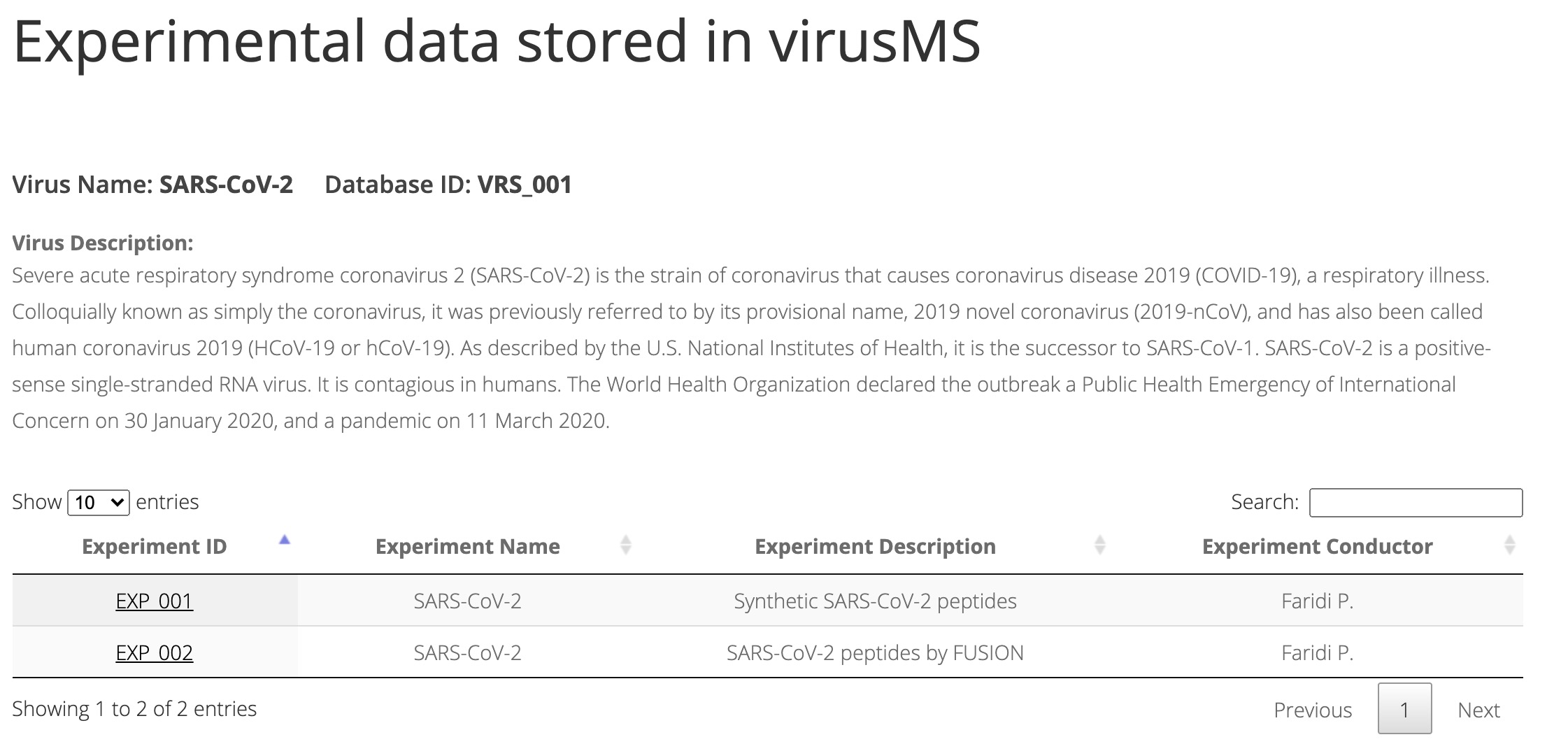
Figure 1. Browsing virusMS database indexed by virus and experiment.
By clicking the specific experiment, the brief information of all peptides related to this experiment will be shown (Figure 2).
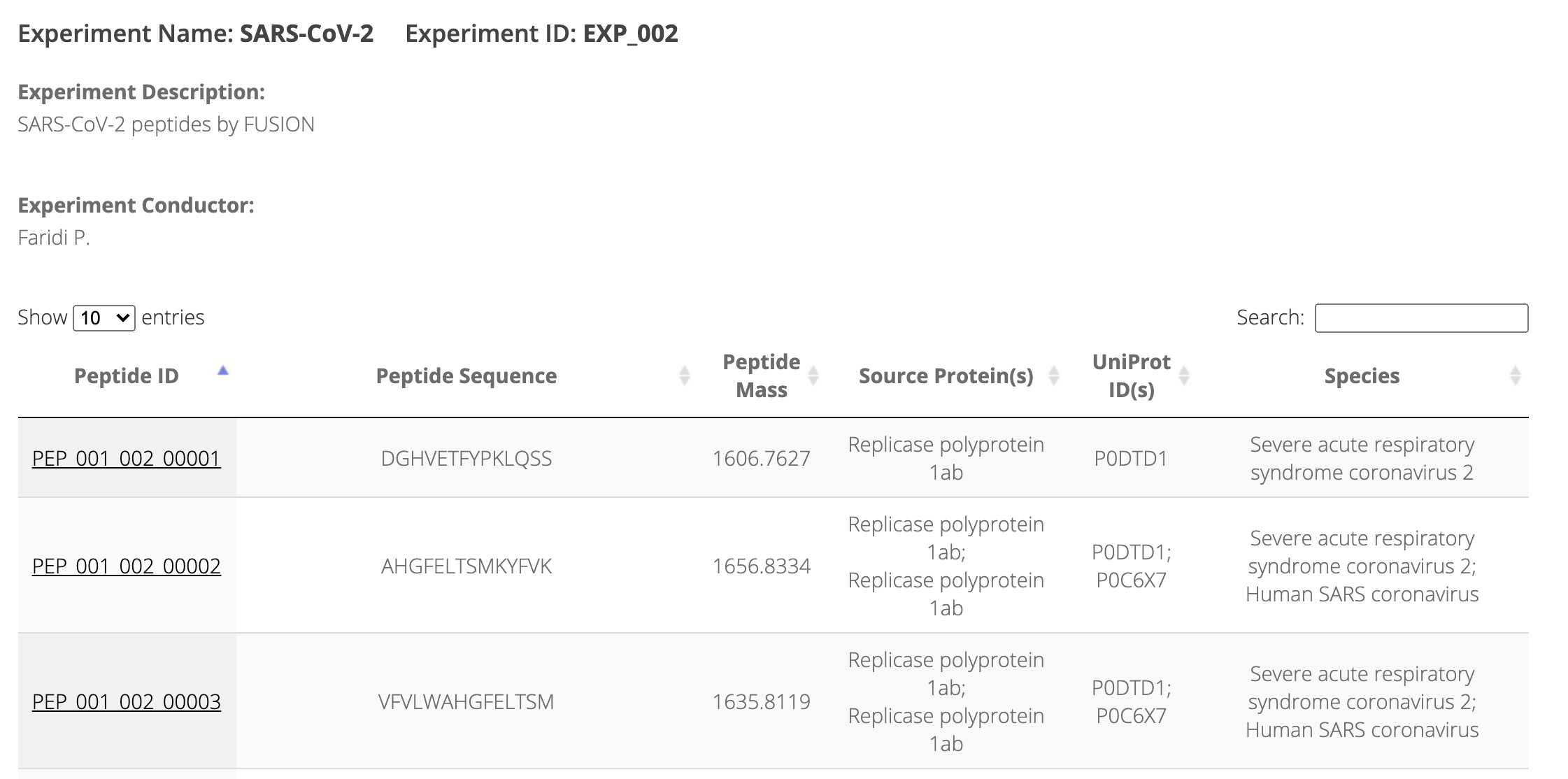
Figure 2. Browsing peptides derived from a specfic experiment.
By clicking the peptide ID (taking PEP_001_001_00004 for example), you will be able to see the detailed information regarding this peptide, including peptide basic information, peptide modification, HLA binding prediction, and the spectrum information of this peptide (Figures 3,4 and 5).
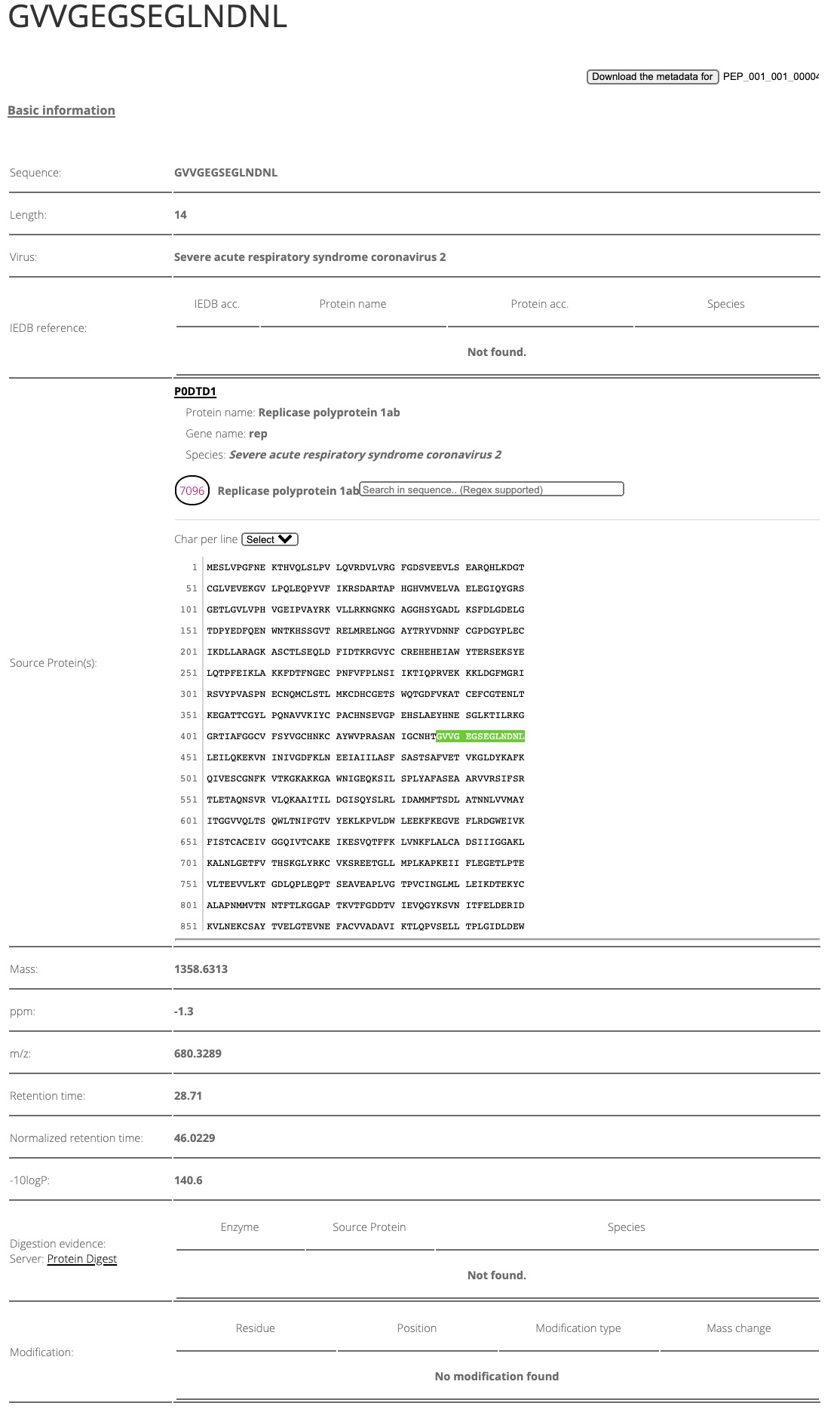
Figure 3. The basic information of the peptide "PEP_001_001_00004".
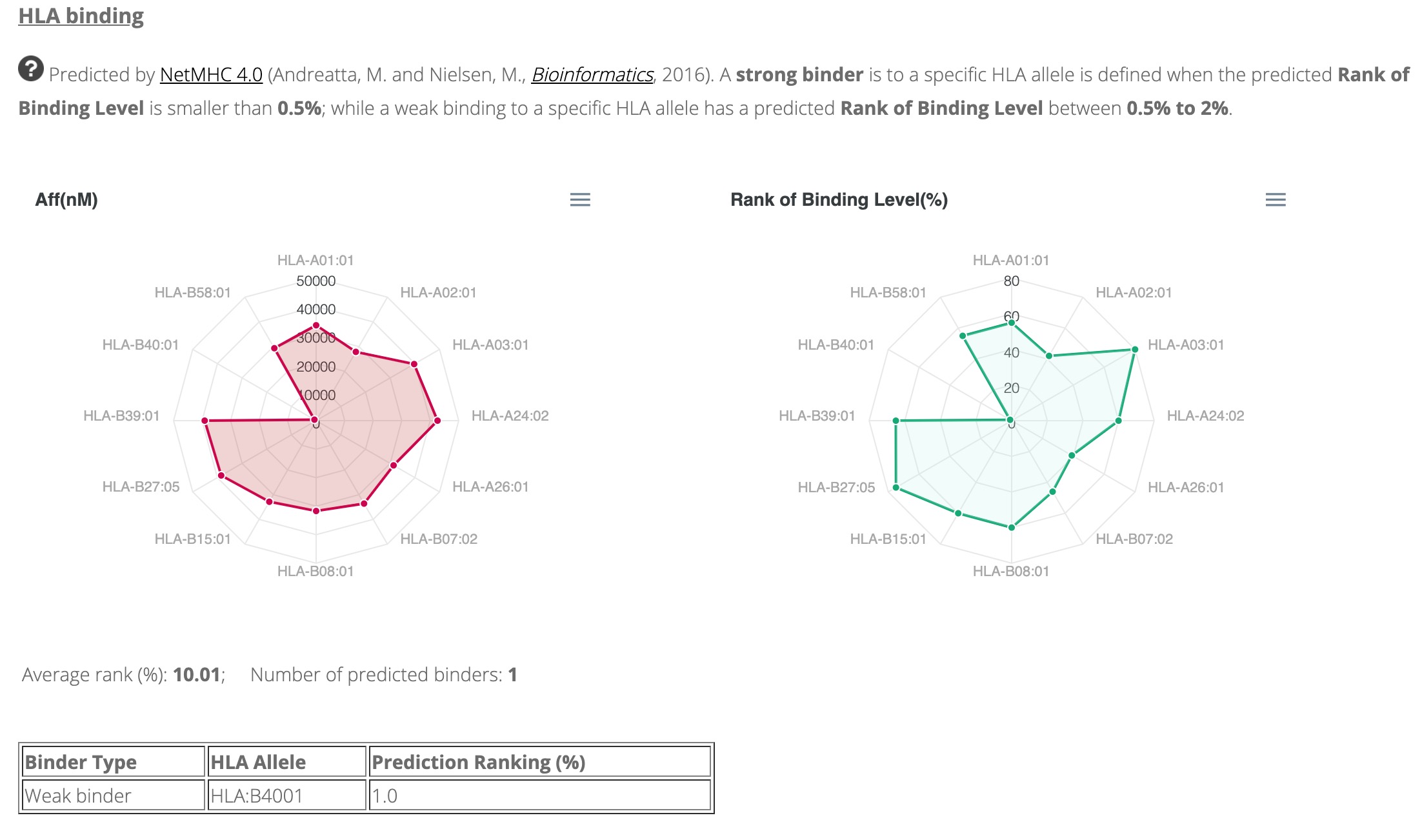
Figure 4. The predicted peptide-HLA-I binding affinities of the peptide "PEP_001_001_00004".
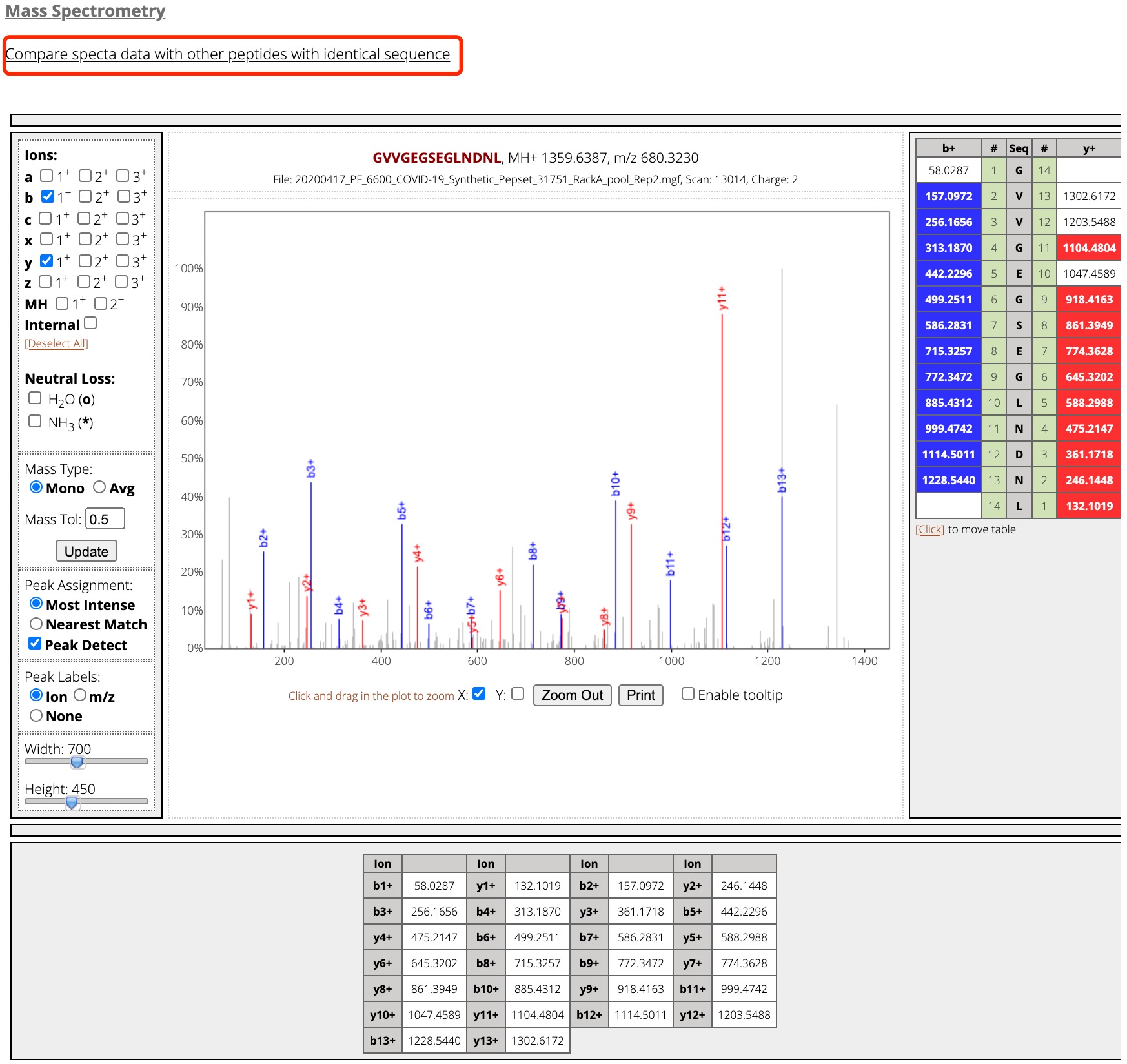
Figure 5. The spectra data of the peptide "PEP_001_001_00004".
By clicking the "Compare specta data with other peptides with identical sequence" link, all spectra data of the peptide with the same sequence but different modifications from two experiments will be compared.
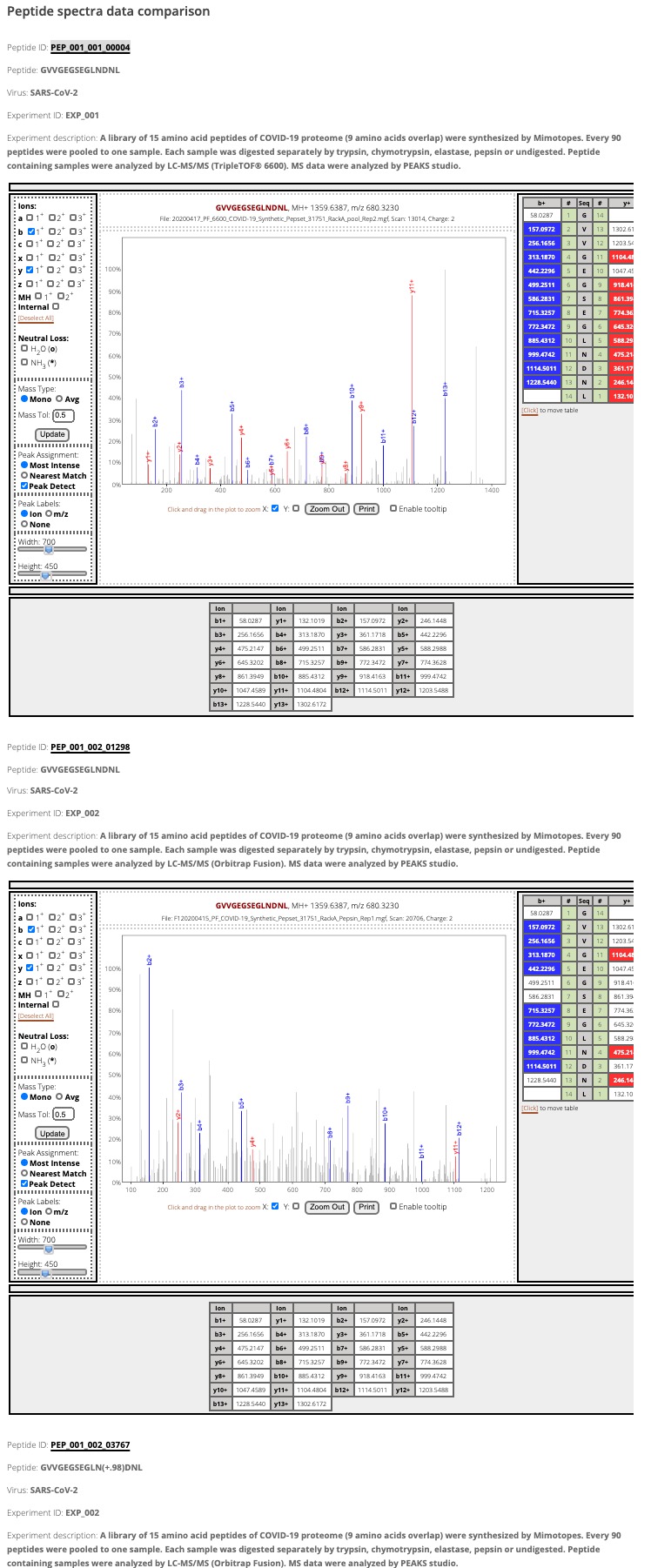
Figure 6. Comapring the spectra data between the peptide "PEP_001_001_00004" and other peptides with the identical sequence.
Searching virusMS is fairely easy. Just go to the search page and search the database using different keywords, as demonstrated in Figure 1.
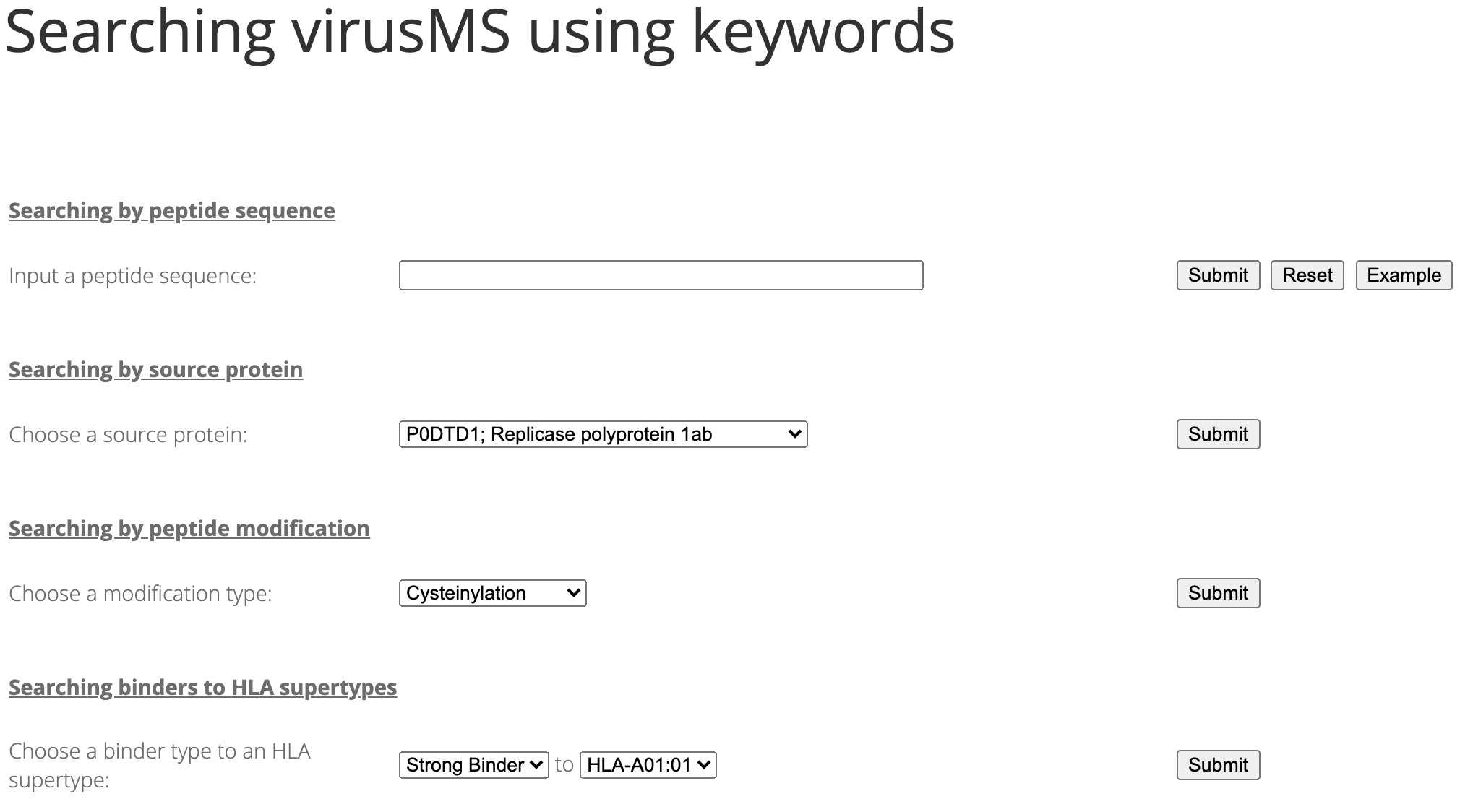
Figure 1. Searching virusMS using different keywords.
By clicking the ID of the interested peptide, the detailed information will be shown in another page (Figure 2).
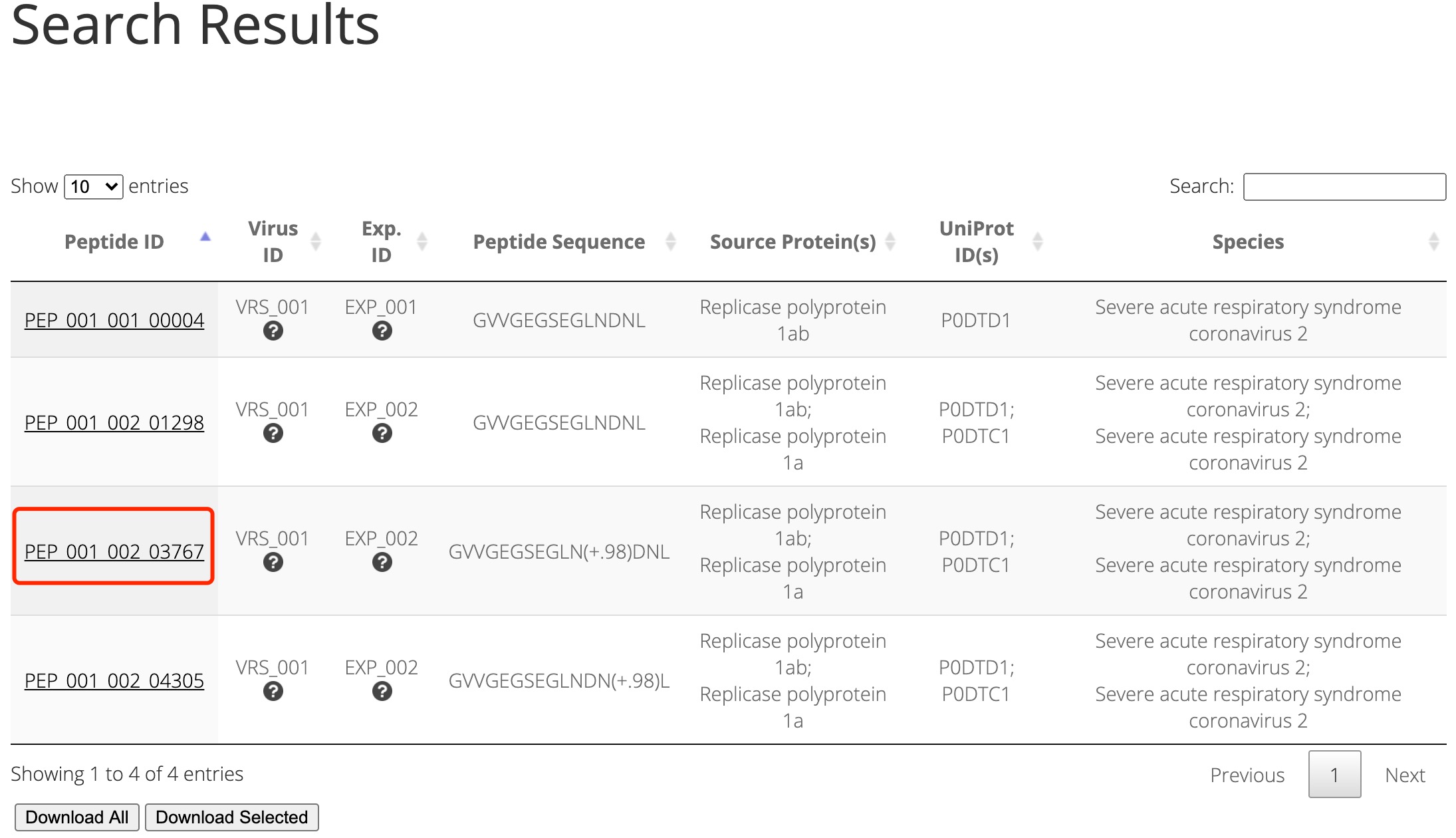
Figure 2. Searching results using 'GVVGEGSEGLNDNL' as a keyword.
If the peptide is not in our experiments, virusMS will instead show entries for this peptide in our proteome predcition results, as demonstrated in Figure 3.
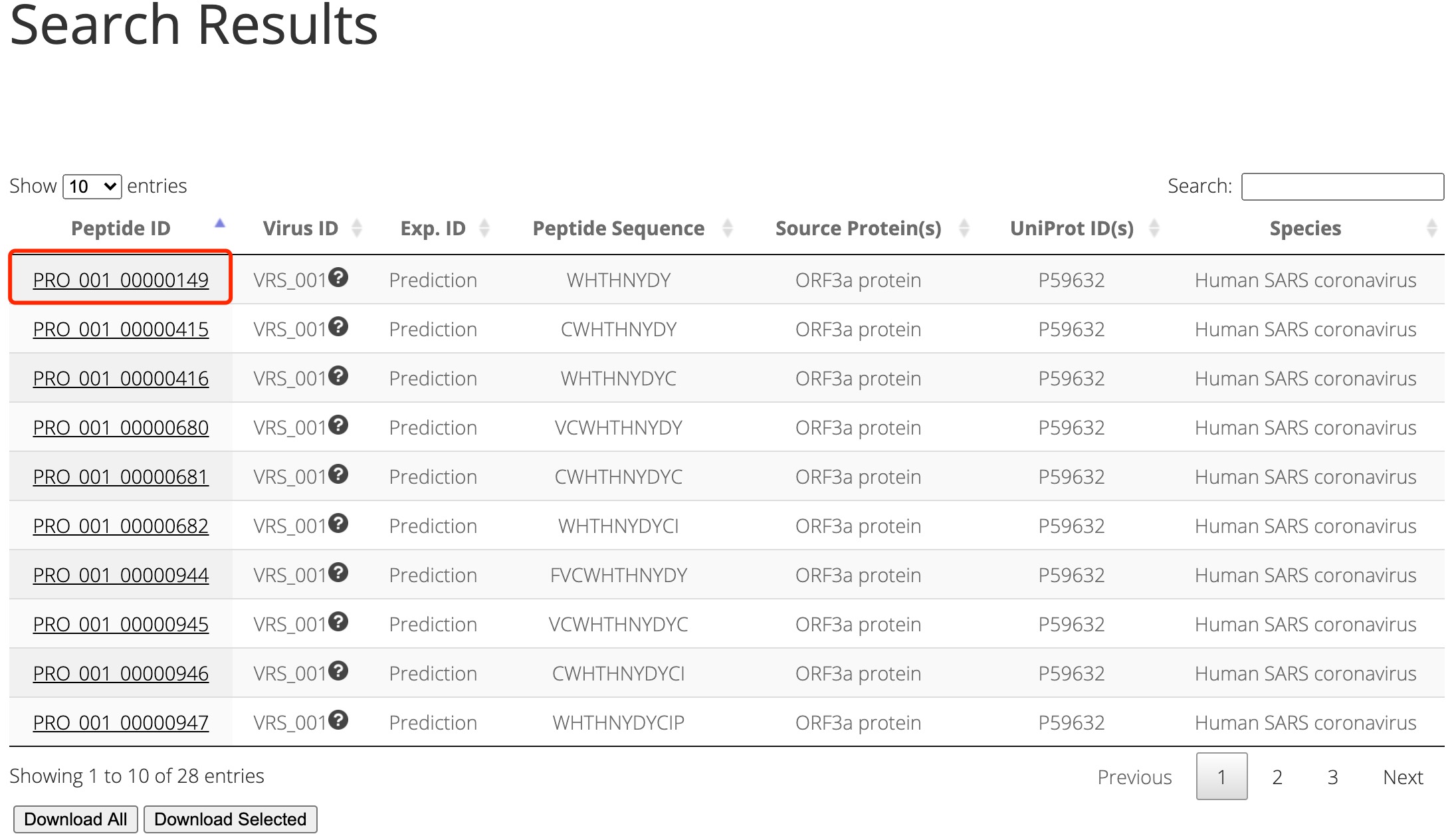
Figure 3. Searching results using 'WHTHNYDY' as the peptide sequence keyword.
By clicking any entry, a webpage will be presented to show very basic informaiton for this peptide. As this peptide is not found in our experiments, only basic identity and prediction information for this peptide will be shown, as demonstrated in Figure 4.
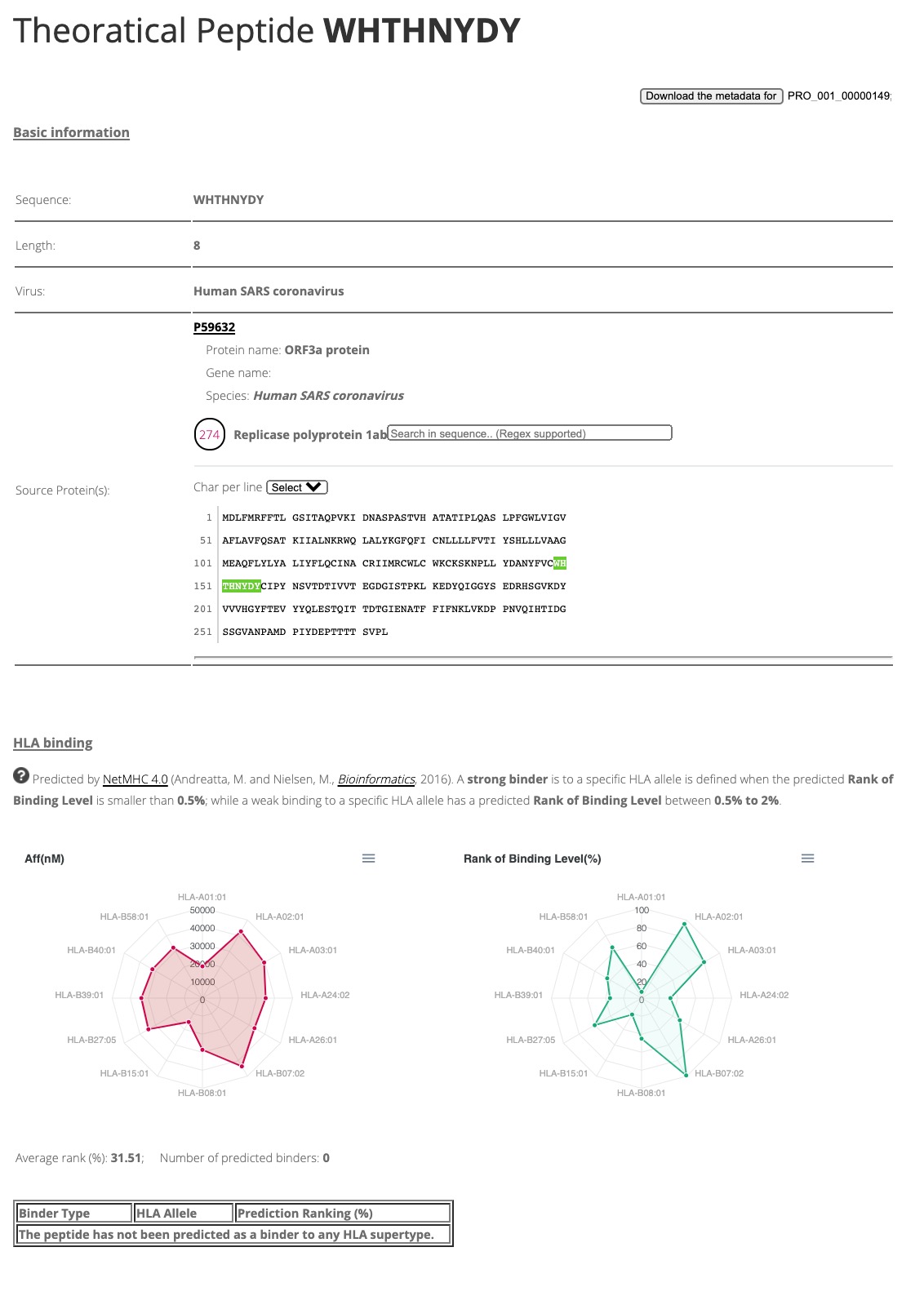
Figure 4. Basic information of theoratical peptide 'WHTHNYDY'.
virusMS is an open-access knowledge base and all information documented in the database is available for download. Please select the following information to download.
Click here to download the database file (.sql);
Click the following links to download all the peptide entries and their information from virusMS (in .csv format);
SARS-CoV-2
Click here to download the spectral data of all the peptides documented in the database.
Click the following links to download the peptide-HLA-I binding prediction results by NetMHC4.0 for the documented peptide in virusMS.
SARS-CoV-2
Click here to download the file index describing our data stored in the PRIDE repository (PXD022191).
In addition, users can select the peptides of interest to download by searching. For example, when using the peptide sequence keyword 'GVVGEGSEGLNDNL'. The search results is shown as follows and users can choose any row(s) (i.e. peptide(s)) they would like to download. Then click the 'Download selected' button to download. Users can also click the 'Download all' button to download all the peptides extracted from virusMS (Figure 1).
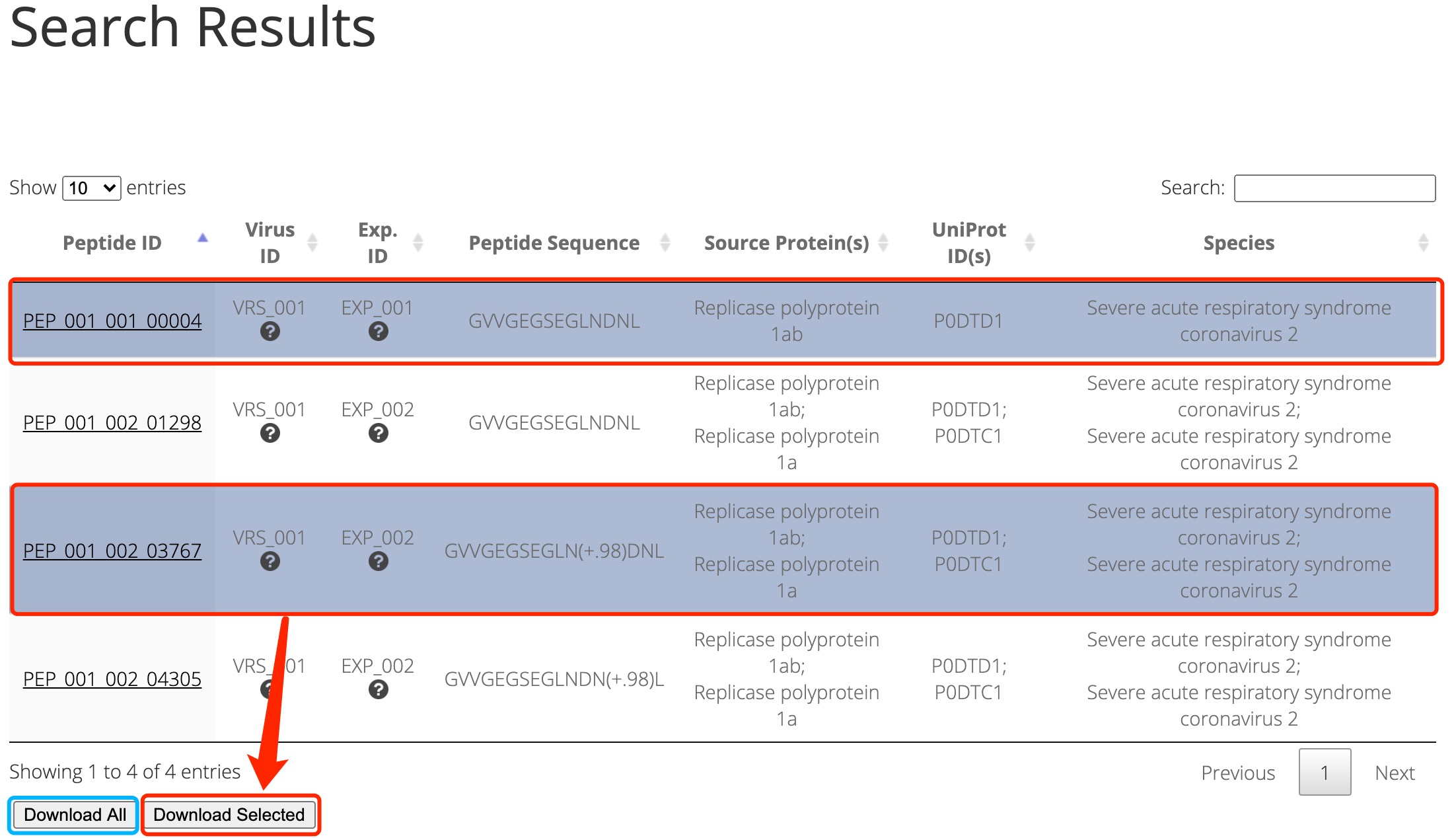
Figure 1. Downloading peptides from searching results.
Alternatively, users can click to view one peptide and then download the meta-data on the detailed information page, by clicking the 'Download the meta-data' button (Figure 2).

Figure 2. Downloading the peptide information on the peptide detailed information page.